当生物与Python交织之时
故事将就此展开
接触 计算机技术(IT) 也已经有些年头了
计算机技术表现出来的 高效 的生产力着实让人赞叹, 让人有无穷的动力参与其中
如今入了生物的坑, 自然一直有探索各种生物和信息技术交融的知识领域
但是浅尝辄止的兴趣式的游玩, 终究是不太够用的
倒不如静下心来, 实际地系统地学习一些内容
⚓️书籍简介 📚
挑选的书是从 江科大长山校区图书馆 里翻找到的
位于 自然科学图书第2阅览室(5楼), 5号架B面5层, 索书号: Q811.4/51
@信息来自江科大图书馆v5.6书目检索系统
题名/责任者: Python生物信息学数据管理/(意) Allegra Via, (德) Kristian Rother, (意) Anna Tramontano著 卢宏超, 陈一情, 李绍娟译
出版发行项: 北京:电子工业出版社,2017.01
ISBN及定价: 978-7-121-30382-1/CNY69.00
载体形态项: 17, 318页:图;26cm
中图法分类号:Q811.4-39
版本附注:由Taylor & Francis出版集团旗下,CRC出版公司授权翻译出版
提要文摘附注:
本书由六部分组成:Python语言基本介绍,语言所有成分介绍,高级编程,数据可视化,生物信息通用包Biopython,最后给出20个"编程秘笈”,范围涵盖了从二级结构预测、多序列比对到蛋白质三维结构的广泛话题。此外,附录还包括了大量的生物信息常用资源的信息。
不过找也费了番功夫, 而且发现学校关于生物信息学方面的书还真的不是非常多
可见生物信息学的确还是比较新的一门学科
⚓️写在前面 ✍
此读书笔记会忽略亦或者增加一些内容, 比如
- 忽略基础的
Python语法: 本书还是使用了大量的例子来讲解Python的基础语法的, 但是对于笔者来说已无必要. 需要补充基础语法的小伙伴可以看这里 => Python3 教程 | 菜鸟教程 (runoob.com) - 增加一些涉及到的生物学理论细节: 这主要是补充书中的内容, 使得易于理解相关的例子
当然, 其实主要是什么值得补充就会补充的 :happy:
本笔记按照原书的目录顺序展开, 但是仅保留少部分的标题, 主要以技术流程来整合笔记
本书的代码使用的Python的版本为 Python 2.6.6
笔者使用由 Anaconda 构建的Python版本为 python-2.7.13 的虚拟环境来复现书中的代码
期间可能会尝试使用 Python3.8 的版本来重构里面的代码
没有特别说明, 笔记里的Python版本则为 python-2.7.13
事实上, 掌握充足的生物学知识是非常重要的
计算机技术只是帮助我们自动化地完成了基于生物学认识的工作
本书也只是讲解了如何将Python引入分析以及数据管理的工作流
当你掌握真正的算法和理论实验背景, 你将可以更好地利用Python这个强大的工具
多多做实验, 看看理论书吧
⚓️故事就此展开 📖
⚓️第一部分 入门
本书的作者描述了他希望这本书带给读者什么
我们希望本书能成为帮助读者每天攀登数据之山的地图, 希望读者能让编程为工作提供便利, 而不必先成为一个专业的软件开发者
作者建议像学习德语一样, 学习Python语法, 把Python标准库 当作字典来查
Python标准库 :
书中的代码和一些总结参考:
⚓️第1章 Python shell
关于Python的安装可以参考这篇文章 Anaconda之路
主要介绍了如何使用Anaconda来构建Python虚拟环境
Anaconda是一个可以管理Python包和Python环境的软件所谓
Python包指的是别人写(第三方)的有一定功能的Python代码所谓
Python环境其实就是Python代码运行时调用的具体版本的Python解释器所谓
管理指的就是我们可以通过Anaconda的conda指令, 随意地创建包含有我们指定的Python包的指定版本的Python环境, 这样做的目的是解决兼容性问题(相互依赖问题)比如这本书的代码是
Python2时代编写的, 我自然是不可以用Python3的解释器来运行的.那就可以创建一个
Python2的环境来复现书中的代码, 然后再创建一个Python3的环境来重新编写他们(重构)
这样我们就可以复现书中的代码了, 而且还可以轻松的重构老旧的代码
⚓️1.2 案例: 计算ATP水解的ΔG
吉布斯自由能:
在热力学里,吉布斯能(英语:Gibbs Free Energy),又称吉布斯自由能、吉布斯函数、自由焓,常用英文字母
标记。吉布斯能是国际化学联会建议采用的名称。吉布斯能是描述系统的热力性质的一种热力势,定义为[1]:101[2]:128-129
$$
\Delta G=\Delta G^{0}+R T * \ln \left([A D P] *\left[P_{i}\right] /[A T P]\right)
$$
$$
[?]: 指 ? 的摩尔浓度 \\ ΔG : 吉布斯自由能 \\ ΔG^0 : 标准反应吉布斯能 \\ R : 摩尔气体常数 \\ T : 温度 \\
$$
- python的
math模块:
1 | import math # 导入数学计算模块 |
⚓️1.3.4 计算
这里引入一篇博文来补充各种符号的意思
特别之处在Python中, ^ 符号
| 符号 | 功能 | 例子 |
|---|---|---|
<sup> |
按位异或运算符:当两对应的二进位相异时,结果为1 | (a b) 输出结果 49 ,二进制解释: 0011 0001 |
** |
幂 - 返回x的y次幂 | a**b 为10的20次方, 输出结果 100000000000000000000 |
参见 Python2.7.18文档 @9.9.1. 将运算符映射到函数
- 计算流程:
1 | ATP = 3.5 |
通过导入各种参数, 可以直接计算出吉布斯自由能
⚓️第2章 第一个Python程序
⚓️2.2案例: 如何计算胰岛素序列中的氨基酸频率 *
- 计算流程:
1 | # insulin [Homo sapiens] GI:386828 |
-
“字符串1”.count(“字符串2”) : 返回
字符串1中含有多少字符串2的数量-
string.``count(s, sub[, start[, end]])Return the number of (non-overlapping) occurrences of substring sub in string
s[start:end]. Defaults for start and end and interpretation of negative values are the same as for slices.
-
注意缩进错误:IndentationError: unindent does not match any outer indentation level
切不可混用 空格缩进和制表符缩进, 否则你会疯的
- Python 的反斜杠换行:
1 | 3 + 5 + \ |
-
内建函数(build-in): 2. 内置函数 — Python 2.7.18 文档
比如
len()可以直接调用, 并返回结果 -
对象方法: 某个对象(类的实例)的内部的方法(函数), 通过
.符号调用, 如" ".count(' ') -
in迭代:
1 | for ... in ...: |
补充 in
,对print输出的影响:
1 | print 1,2, # 注意末尾的逗号可以抑制换行符 |
-
转义字符
\: -
引号之间的嵌套:
-
random模块: 9.6. random — 生成伪随机数 — Python 2.7.18 文档
⚓️2.5自测题
2009年, Elizabeth H. Blackburn, Carol W. Greide 和 Jack W. Szostak 因发现端粒酶功能而获得诺贝尔奖, 端粒酶负责延伸染色体的末端. 通过NCBI的蛋白质数据库检索人类亚型I的端粒的11132个残基序列, 试问其中哪种氨基酸出现的最为频繁?
⚓️第二部分 数据管理
⚓️第3章 分析数据列
译者推荐标准库模块 collections :
8.3. collections — High-performance container datatypes — Python 2.7.18 文档
⚓️3.2案例: 树突长度
作者使用了 ImageJ 来计算树突的长度并把值传入文本文件
软件指路: ImageJ
in迭代:
1 | # for循环中按行迭代文件的方法 |
- 内建函数(内置函数): 2. 内置函数 — Python 2.7.18 文档
1 | # 内建函数 |
-
%格式化输出:: -
文件的
readlines方法: file.readlines([sizehint]) — Python 2.7.18 文档
此方法可以将文件的内容按行组合成列表返回
-
硬编码(hard-coding): -
不关闭文件的隐患:
当在其他时候读取文件的时候, 之前写入的部分内容因为没有超过缓冲区大小, 还处于内存内.
这将导致有时候你将无法获取想要的内容, 切记及时关闭文件.
-
缓冲区: -
文件的
writelines方法:
将列表按项无空格连续写入文件, 列表内只支持 字符串 .
file.writelines(sequence)¶Write a sequence of strings to the file. The sequence can be any iterable object producing strings, typically a list of strings. There is no return value. (The name is intended to match
readlines();writelines()does not add line separators.)
" ".join()方法:
返回一个由可迭代对象中的字符串串联" "组成的字符串。
str.join(iterable)¶
Return a string which is the concatenation of the strings in iterable. If there is any Unicode object in iterable, return a Unicode instead. A
TypeErrorwill be raised if there are any non-string or non Unicode object values in iterable. The separator between elements is the string providing this method.
⚓️第4章 解析数据记录
⚓️4.2 案例: 整合质谱数据, 转化到代谢通路中
-
UniProt形式: UniProtThe mission of UniProt is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information.
- ~ AC:
-
CSV逗号分隔文件: -
TSV制表符分隔文件: -
MS质谱: -
作者提到的软件与网站:
-
Mascot search engine: Protein identification software for mass spec(质谱) data
Welcome to the home of Mascot software, the benchmark for identification, characterisation and quantitation of proteins using mass spectrometry data. Here, you can learn more about the tools developed by Matrix Science to get the best out of your data, whatever your chosen instrument.
-
Reactome Pathway Database:
代谢通路资源获取Reactome is a free, open-source, curated and peer-reviewed pathway database. Our goal is to provide intuitive bioinformatics tools for the visualization, interpretation and analysis of pathway knowledge to support basic research, genome analysis, modeling, systems biology and education.
If you use Reactome in Asia, we suggest using our Chinese mirror site at reactome.ncpsb.org.
-
1 | # proteins participating in cell cycle 参与细胞通路的蛋白 |
1 | # if 语句 |
- 列表推导式: 5.1.4. 列表推导式
1 | bases = ['A','C','T','G'] |
-
ESR1蛋白:Protein : Estrogen receptor 雌激素受体
Gene : ESR1
Organism : Homo sapiens (Human)
-
蛋白详情ESR1 - Estrogen receptor - Homo sapiens (Human) - ESR1 gene & protein (uniprot.org)
-
ESR1蛋白序列: https://www.uniprot.org/uniprot/P03372.fasta>sp|P03372|ESR1_HUMAN Estrogen receptor OS=Homo sapiens OX=9606 GN=ESR1 PE=1 SV=2
MTMTLHTKASGMALLHQIQGNELEPLNRPQLKIPLERPLGEVYLDSSKPAVYNYPEGAAY
EFNAAAAANAQVYGQTGLPYGPGSEAAAFGSNGLGGFPPLNSVSPSPLMLLHPPPQLSPF
LQPHGQQVPYYLENEPSGYTVREAGPPAFYRPNSDNRRQGGRERLASTNDKGSMAMESAK
ETRYCAVCNDYASGYHYGVWSCEGCKAFFKRSIQGHNDYMCPATNQCTIDKNRRKSCQAC
RLRKCYEVGMMKGGIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAANLWPSPLMIKR
SKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINW
AKRVPGFVDLTLHDQVHLLECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEG
MVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSLEEKDHIHRVLD
KITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLL
LEMLDAHRLHAPTSRGGASVEETDQSHLATAGSTSSHSLQKYYITGEAEGFPATV
-
1 | # 读取整个FASTA记录, 找到里面所属为'Homo sapiens'的序列 |
⚓️第5章 搜索数据
⚓️5.2案例: 将RNA序列翻译为相应的蛋白质序列
1 | codon_table={ |
-
str.startswith(prefix[, start[, end]]) 5. 内置类型 — Python 2.7.18 文档如果字符串以指定的 prefix 开始则返回
True,否则返回False。 prefix 也可以为由多个供查找的前缀构成的元组。 如果有可选项 start,将从所指定位置开始检查。 如果有可选项 end,将在所指定位置停止比较。 -
核酸序列
(开放)阅读框(Open Reading Frame):-
由于核糖体翻译mRNA时, 三个碱基翻译为一个氨基酸, 那么同一个序列理论上存在三个不同的阅读框
-
对应于
移码突变对于 序列
ATCGATCG有ATC GAT CG,A TCG ATC G,AT CGA TCG三种方法解读 -
ORF指的是DNA上的序列,从5’端翻译起始密码子ATG到终止密码子(TAA,TAG,TGA)的蛋白质编码序列。对于任意给定的一段DNA,有两个问题需要考虑,-
一是DNA双链中的哪条是编码链
-
二是编码区究竟从第一个碱基开始进行编码
所以每条链都有潜在的3种ORF,而对于双链DNA来说就有6种可能的ORF。也就是说先从给定的DNA单链为模版,分别从5’-3’方向第123个碱基开始翻译,再以互补链为模版,分别从3’-5’方向的第123个碱基开始翻译,得到另外3种翻译结果。正链上的正向读码框forward,负链的为反向读码框reverse。6个潜在ORF中,一般选择中间没有被终止密码子隔开的最大的读码框为正确的结果。
-
-
-
文件读取的
.next()方法: file.next 2.7文档file.next()A file object is its own iterator, for example
iter(f)returns f (unless f is closed). When a file is used as an iterator, typically in aforloop (for example,for line in f: print line.strip()), thenext()method is called repeatedly. This method returns the next input line, or raisesStopIterationwhen EOF is hit when the file is open for reading (behavior is undefined when the file is open for writing). In order to make aforloop the most efficient way of looping over the lines of a file (a very common operation), thenext()method uses a hidden read-ahead buffer. As a consequence of using a read-ahead buffer, combiningnext()with other file methods (likereadline()) does not work right. However, usingseek()to reposition the file to an absolute position will flush the read-ahead buffer. -
for循环中的else: break-and-continue-statements-and-else-clauses-on-loops
1 | for n in range(2, 10): |
- 预测蛋白质序列中的无序(环)区域: 既不是
α螺旋也不是β折叠的区域, 就是无序的.核心思想: 每个氨基酸都有一个特定的二级结构元件倾向, 通过大量已知的蛋白质结构(PDB)的二级结构元件中各类氨基酸类型出现的频率f,来估计倾向.- 使用
1-f计算:f是以有二级结构元件的部分为基数的(就是有序的), 那么在这样的基数里某种氨基酸出现的越多(频数接近1)则倾向于有序的可能性越大. - 对20个氨基酸倾向的值进行标准化和引入负值, 设定一个是否为无序的
阈值, 大于阈值为无序(环), 小于为有序(二级结构).
1 | propensities ={ |
-
-
数据示例概览: RCSB PDB - 6XM5: Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71HEADER VIRAL PROTEIN 29-JUN-20 6XM5
TITLE STRUCTURE OF SARS-COV-2 SPIKE AT PH 5.5, ALL RBDS DOWN
COMPND MOL_ID: 1;
COMPND 2 MOLECULE: SPIKE GLYCOPROTEIN;
...
COMPND 6 MUTATION: YES
SOURCE MOL_ID: 1;
SOURCE 2 ORGANISM_SCIENTIFIC: SEVERE ACUTE RESPIRATORY SYNDROME CORONAVIRUS
...
SOURCE 9 EXPRESSION_SYSTEM_CELL_LINE: FREESTYLE 293F
KEYWDS SARS-COV-2 SPIKE, COVID19, VIRAL PROTEIN
EXPDTA ELECTRON MICROSCOPY
AUTHOR T.ZHOU,Y.TSYBOVSKY,A.OLIA,P.D.KWONG
REVDAT 2 12-AUG-20 6XM5 1 JRNL HETSYN
REVDAT 1 29-JUL-20 6XM5 0
JRNL AUTH T.ZHOU,Y.TSYBOVSKY,A.S.OLIA,J.GORMAN,M.A.RAPP,G.CERUTTI,
...
JRNL TITL CRYO-EM STRUCTURES DELINEATE A PH-DEPENDENT SWITCH THAT ...
JRNL PMID 32637958
JRNL DOI 10.1101/2020.07.04.187989
REMARK 2
...
REMARK 3 SOFTWARE PACKAGES : LATITUDE, CTFFIND, UCSF CHIMERA, COOT,
REMARK 3 PHENIX, RELION, RELION, RELION, RELION
...
REMARK 3 OVERALL ANISOTROPIC B VALUE : NULL
...
REMARK 900 STRUCTURE OF SARS-COV-2 SPIKE AT PH 5.5, ALL RBDS DOWN
DBREF 6XM5 A 14 1208 UNP P0DTC2 SPIKE_SARS2 14 1208
DBREF 6XM5 B 14 1208 UNP P0DTC2 SPIKE_SARS2 14 1208
DBREF 6XM5 C 14 1208 UNP P0DTC2 SPIKE_SARS2 14 1208
SEQADV 6XM5 GLY A 682 UNP P0DTC2 ARG 682 ENGINEERED MUTATION
SEQADV 6XM5 SER A 683 UNP P0DTC2 ARG 683 ENGINEERED MUTATION
...
SEQADV 6XM5 LYS C 1288 UNP P0DTC2 EXPRESSION TAG
SEQRES 1 A 1275 GLN CYS VAL ASN LEU THR THR ARG THR GLN LEU PRO PRO
SEQRES 2 A 1275 ALA TYR THR ASN SER PHE THR ARG GLY VAL TYR TYR PRO
...
SEQRES 99 C 1275 LYS
HET NAG D 1 14
HET NAG D 2 14
...
HETNAM NAG 2-ACETAMIDO-2-DEOXY-BETA-D-GLUCOPYRANOSE
...
FORMUL 4 NAG 58(C8 H15 N O6)
HELIX 1 AA1 ASP A 294 LYS A 304 1 11
...
SHEET 3 AA1 8 TYR A 265 TYR A 269 -1 O TYR A 265 N PHE A 65
...
LINK O4 NAG S 1 C1 NAG S 2 1555 1555 1.44
CRYST1 1.000 1.000 1.000 90.00 90.00 90.00 P 1
ORIGX1 1.000000 0.000000 0.000000 0.00000
...
SCALE3 0.000000 0.000000 1.000000 0.00000
ATOM 1 N ALA A 27 188.584 130.357 168.091 1.00 91.79 N
ATOM 2 CA ALA A 27 189.571 130.261 169.159 1.00 91.79 C
ATOM 3 C ALA A 27 190.053 131.641 169.567 1.00 91.79 C
ATOM 8249 OG SER A1147 191.132 184.224 284.141 1.00152.29 O
TER 8250 SER A1147
ATOM 8251 N ALA B 27 132.579 208.265 174.154 1.00140.96 N
...
TER 24584 SER C1147
HETATM24585 C1 NAG D 1 184.739 152.280 150.742 1.00114.03 C
HETATM24586 C2 NAG D 1 183.632 153.136 150.126 1.00114.03 C
...
CONECT 29225033
CONECT 70225047
...
CONECT2539625389
MASTER 907 0 58 69 173 0 0 625393 3 936 297
END
-
⚓️5.5 自测题
-
NCBI下载FASTA格式RNA序列: -
PubMed文献检索: -
二级结构元素预测:
- 氨基酸二级结构偏好表: Amino Acid Secondary Structure Preferences
| A.A. | Helix | Helix | Sheet | Sheet |
|---|---|---|---|---|
| A.A. | Designation | P | Designation | P |
| A | H | 1.45 | I | 0.97 |
| C | i | 0.77 | h | 1.30 |
| D | i | 0.98 | i | 0.80 |
| E | H | 1.53 | B | 0.26 |
| F | h | 1.12 | h | 1.28 |
| G | B | 0.53 | i | 0.81 |
| H | h | 1.24 | b | 0.71 |
| I | I | 1.00 | H | 1.60 |
| K | I | 1.07 | b | 0.74 |
| L | H | 1.34 | h | 1.22 |
| M | h | 1.20 | H | 1.67 |
| N | b | 0.73 | b | 0.65 |
| P | B | 0.59 | b | 0.62 |
| Q | h | 1.17 | h | 1.23 |
| R | i | 0.79 | i | 0.90 |
| S | i | 0.79 | b | 0.72 |
| T | i | 0.82 | h | 1.20 |
| V | h | 1.14 | H | 1.65 |
| W | h | 1.14 | h | 1.19 |
| Y | b | 0.61 | h | 1.29 |
-
Designations:- H = Strong Former, h = Former, I = Weak Former, i = Indifferent, B = Strong Breaker, b = Breaker;
-
P: Conformational Parameter
⚓️第6章 过滤数据
⚓️6.2 案例: 使用RNAseq输出数据
-
新一代测序(Next-generation sequencing,NGS):
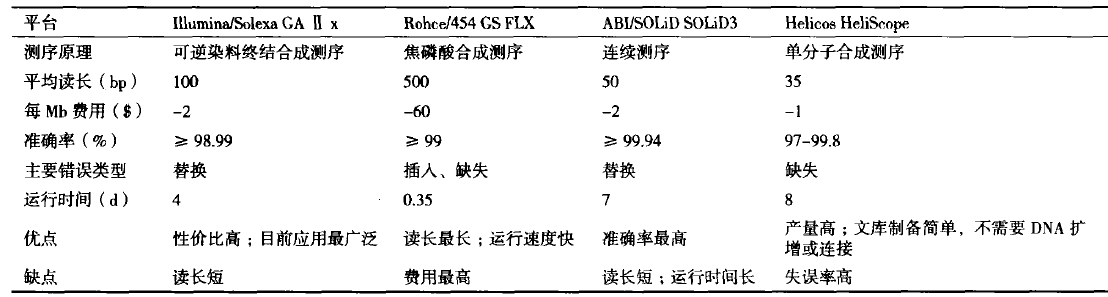
最早广泛应用测序技术为 70 年代的 Sanger 法,这也是完成人类基因组计划的基础,因其测序通量低、费时费力,科学家们一直在寻求通量更高、速度更快、价格更便宜、自动化程度更高的测序技术。自 2005 年以来,以Roche 公司的 454 技术、Illumina 公司的 Solexa 技术以及 ABI 公司的 SOLiD 技术为标志的高通量测序技术相继诞生 [6]。相较于传统方法,该技术主要特点是测序通量高、测序时间和成本显著下降,可以一次对几十万到几百万条 DNA 分子序列测定,这使某物种全基因组和转录组的全貌细致分析成为可能,又称为深度测序,很多文献中称其为新一代测序技术,足见其划时代意义 [7]。
-
转录组(Transcriptome)测序技术|RNA-Seq(RNA sequencing)技术 :
-
转录本就是基因通过转录形成的通过剪切生成的一种或多种可供编码蛋白质的成熟的mRNA。剪切的过程中可能会剪切掉外显子,也有可能保留部分内含子,这样就形成了多种mRNA即多个转录本。
@基因研究不知道转录本,做什么引物设计?! (sohu.com) -
RNA-Seq利用高通量测序技术对组织或细胞中所有RNA 反转录而成cDNA文库进行测序,通过统计相关读段(reads)数计算出不同RNA的表达量,发现新的转录本;如果有基因组参考序列,可以把转录本映射回基因组,确定转录本位置、剪切情况等更为全面的遗传信息,已广泛应用于生物学研究、医学研究、临床研究和药物研发等。
@转录组测序技术的应用及发展综述 - 知乎 (zhihu.com) 推荐阅读 -
大致流程:
比对read到基因组,转录本组装,差异表达分析和差异基因注释
转录本组装主要就是将比对到参考基因组的read组装成转录本接着进行下游的新转录本鉴定和转录本定量等分析,常见软件有StringTie和cufflinks。而对于无参转录组分析来说,转录本组装主要是将测序reads从头组装成转录本,常见软件有Trinity,Oases和SOAPdenovo-Trans。
-
无参转录组和有参转录组的区别: 如果所研究的物种有组装注释质量较好基因组序列,且和该基因组序列比对效率较高,那么可以采用有参转录组的分析策略,直接进行分析。反之,则需要按照无参转录组的分析策略进行转录本组装,构建unigene(基因簇)库,然后进行后续分析。
@转录组测序:常见基础知识准备一 - 知乎 (zhihu.com)
-
-
转录本文件
.gtf: 记录了各个样本装配好的转录本 -
Cufflinks (cole-trapnell-lab.github.io)
Cufflinks assembles transcripts, estimates their abundances, and tests for differential expression and regulation in RNA-Seq samples. It accepts aligned RNA-Seq reads and assembles the alignments into a parsimonious set of transcripts. Cufflinks then estimates the relative abundances of these transcripts based on how many reads support each one, taking into account biases in library preparation protocols.
Cufflinks includes a program that you can use to help analyze the transfrags you assemble. The program cuffcompare helps you:
- Compare your assembled transcripts to a reference annotation
- Track Cufflinks transcripts across multiple experiments (e.g. across a time course)
-
转录本比对文件
.tracking: 导入转录本文件(.gtf)到cuffcompare生成 -
元组方法:
-
intersection(*others) : 返回一个新集合,其中包含原集合以及 others 指定的所有集合中共有的元素。 -
difference(*others) : 返回一个新集合,其中包含原集合中在 others 指定的其他集合中不存在的元素。 -
更多方法参见文档
-
-
列表的
.count()方法: 8.6. array — 高效的数值数组 — Python 2.7.18 文档array.count(x) : 返回 x 在数组中的出现次数。
-
内置函数相关: 2. 内置函数 — Python 2.7.18 文档
pop():reduce():del():remove():enumerate():
-
.readlines()返回的列表可以索引:1
2lines = open('text.txt').readlines()
open('new.txt', 'w').writelines(lines[2:4]+lines[6:]) -
使用
列表推导式筛选:1
2data_a = [1, 2, 3, 4, 5, 6]
data = [x for x in data_a if x != 3] -
去重妙法-
元组去重:1
2input_file = open('UniprotID.txt')
unique = set(input_file) -
CD-HIT(可容错的高同源性聚类数据库): 可以快速地的基于用户定义的相似性阈值对蛋白质序列进行聚类, 需要输入一组FASTA格式的序列,并返回两个文件: 一个是聚类列表, 一个是所聚各类的代表序列.@CD-HIT Official Website (weizhongli-lab.org)
@weizhongli/cdhit: Automatically exported from code.google.com/p/cdhit (github.com)
⚓️第7章 管理表数据
⚓️7.2 案例: 确定蛋白浓度
-
苯酚福林试剂测定蛋白质含量的流程:-
相关论文: PROTEIN MEASUREMENT WITH THE FOLIN PHENOL REAGENT (jbc.org)
-
对比于
凯氏定氮法的优势,对多个样品可以使用光度计快速地进行测量.
-
-
zip()内置函数:zip([iterable, …]) 2. 内置函数 — Python 2.7.18 文档1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38x = [1, 2, 3]
y = [4, 5, 6]
zipped = zip(x, y)
zipped
[(1, 4), (2, 5), (3, 6)]
x2, y2 = zip(*zipped) # *号的功能是去除zipped元组的外括号(相当于拆解)
x == list(x2) and y == list(y2)
True
# 效果同转置矩阵:
zipped = zip(*zipped)
# 样例
table = [
['protein', 'ext1', 'ext2', 'ext3'],
[0.16, 0.038, 0.044, 0.040],
[0.33, 0.089, 0.095, 0.091],
[0.66, 0.184, 0.191, 0.191],
[1.00, 0.280, 0.292, 0.283],
[1.32, 0.365, 0.367, 0.365],
[1.66, 0.441, 0.443, 0.444]
]
# remove the first row
table = table[1:]
protein, ext1, ext2, ext3 = zip(*table)
# create a single column for `ext` or concatenate each `ext`
# extend (`* 3`) the `protein` column to match the `ext` column.
extinction = ext1 + ext2 + ext3
protein = protein * 3
# create four tuples for each column
table = zip(protein, extinction)
for prot, ext in table:
print prot, ext -
列表创建注意: 如何创建多维列表?编程常见问题 — Python 2.7.18 文档
[0]*3这样的语法是创建了[0]的三个引用, 改变一个列表的内容将改变所有内容
1 | table = [ |
⚓️7.5自测题
-
RNA碱基相似性矩阵:
A G C U A 1.0 0.5 0.0 0.0 G 0.5 1.0 0.0 0.0 C 0.0 0.0 1.0 0.5 U 0.0 0.0 0.5 1.0
⚓️第8章 数据排序
⚓️8.2案例: 数据表排序
-
sorted(iterable[, cmp[, key[, reverse]]]) 内置函数: sorted 2. 内置函数 — Python 2.7.18 文档-
根据 iterable 中的项返回一个新的已排序列表。
-
参数
cmp(x, y): 2. 内置函数 — Python 2.7.18 文档sorted方法会隐式的调用cmp()函数进行两两比对, 可以通过自定义函数传给cmp参数控制排序
Compare the two objects x and y and return an integer according to the outcome. The return value is negative if
x < y, zero ifx == yand strictly positive ifx > y.1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16def numeric_compare(x, y):
return x - y
sorted([5, 2, 4, 1, 3], cmp=numeric_compare)
[1, 2, 3, 4, 5]
def my_cmp(a,b): # 这个cmp可以在比对a和b的第x项相等时,寻找x+1项进行比对
x = 0
if a[x] < b[x]: return 1
if a[x] == b[x]:
if a[x+1] < b[x+1]: return 1
if a[x+1] == b[x+1]:
if a[x+2] < b[x+2]: return 1
if a[x+2] == b[x+2]: return 0
if a[x+2] > b[x+2]: return -1
if a[x+1] > b[x+1]: return -1
if a[x] > b[x]: return -1 -
排序默认按照
ASCII顺序进行排序space 0 : A Q [ a q } ! 1 ; B R \ b r tick “ 2 < C S ] c s { # 3 = D T ^ d t $ 4 > E U _ e u DEL % 5 ? F V ` f v & 6 @ G W g w ‘ 7 H X h x ( 8 I Y i y ) 9 J Z j z * K k + L l , M m - N n . O o / P p
-
-
openator模块的itemgetter: 9.9. operator — 标准运算符替代函数 — Python 2.7.18 文档1
2
3
4
5
6
7inventory = [('apple', 3), ('banana', 2), ('pear', 5), ('orange', 1)]
getcount = itemgetter(1) # 构建一个获取`索引1的值`的itemgetter函数
map(getcount, inventory) # 将inventory按项逐个传入getcount, 获取每项索引号为1的值
[3, 2, 5, 1]
sorted(inventory, key=getcount) # 也可以将getcount作为key, 使用sorted排序
# 这里按照inventory的每一项的索引号为1的值进行排序
[('orange', 1), ('banana', 2), ('apple', 3), ('pear', 5)] -
更多排序参见: 排序指南 — Python 2.7.18 文档
-
通过
shell终端命令排序: Linux sort 命令详解:对文本文件中所有行进行排序。 - Linux 命令搜索引擎 (wangchujiang.com)1
2
3sort myfile.txt # 按照字母表顺序排序
sort -n myfile.txt # 按照数值排序
sort myfile.txt -k2n -kl # 多列排序 -
对
.tsv制表符分隔符文件的每行,按照七个优先级排序
1 | from operator import itemgetter |
- 对BLAST的
.csv逗号分隔符文件每行的某一项进行排序
1 | from operator import itemgetter |
-
blastp(蛋白质比对蛋白数据库)工具: -
A链血红蛋白PDB表报告:
⚓️第9章 模式匹配和文本挖掘
⚓️9.2 案例: 在蛋白质序列中搜索磷酸化模体
-
正则表达式(regular expression, regexp): 代表一组字符串的字符串语法-
类似的: 生物学家使用
N来代替任意碱基(ATCG) -
[和]被称为元字符,由它和字符来编码一组字符串的表达,为正则表达式 -
re.compile(pattern, flags=0): Compile a regular expression pattern into a regular expression object, which can be used for matching using itsmatch()andsearch()methods.
参数见上面的文档 -
re.compile()返回RegexpObject对象, 使用该对象的.search()方法将返回Match Object对象, 该对象还可以使用.group()/.span()/.start()/.end()方法. 另外RegexpObject对象还有.findall()/.match()/.finditer()等方法可以用, 详见文档 -
由(和)配合匹配到的字符串可以用group()获取识别到的更小的子组1
2
3
4
5
6
7seq = 'zzabcdzz'
pattern = re.compile('(a(b)c)d')
match = pattern.search(seq)
print match.group(0)
print match.group(1)
print match.group(2)
print match.groups()
-

-
检索匹配指定序列:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20import re # 正则表达式模块
# define a string with occurrences of regex:
seq = 'VSVLTMFRYAGWLDRLYMLVGTQLAAIIHGVALPLMMLI'
# compile a pattern and assign it to a variable
pattern = re.compile('[ST]Q') # 转换字符串为正则表达式对象
# search for the pattern in the string
match = pattern.search(seq)
if match:
# print the first match along the sequence with the group() method
# 4 characters before and after the pattern
print '%10s' %(seq[match.start() - 4:match.end() + 4])
print '%6s' % match.group()
else:
print "no match"
# 结果:
# MLVGTQLAAI
# TQ -
功能性模体表达:
-
ELM: elm.eu.orgThis computational biology resource mainly focuses on annotation and detection of eukaryotic linear motifs (ELMs) by providing both a repository of annotated motif data and an exploratory tool for motif prediction.
ELMs, or short linear motifs (SLiMs), are compact protein interaction sites composed of short stretches of adjacent amino acids. They are enriched in intrinsically disordered regions of the proteome and provide a wide range of functionality to proteins (Davey,2011,Van Roey,2014) They play crucial roles in cell regulation and are also of clinical importance, as aberrant SLiM function has been associated with several diseases and SLiM mimics are often used by pathogens to manipulate their hosts’ cellular machinery (Davey,2011, Uyar,2014) -
PROSITE: ExPASy - PROSITEPROSITE consists of documentation entries describing protein domains, families and functional sites as well as associated patterns and profiles to identify them [More… / References / Commercial users ].
PROSITE is complemented by ProRule , a collection of rules based on profiles and patterns, which increases the discriminatory power of profiles and patterns by providing additional information about functionally and/or structurally critical amino acids [More…].
-
-
.findall()和.finditer()方法:
1 | import re |
子组group()的使用
1 | import re |
- Linux命令-
grep: Linux grep 命令详解:强大的文本搜索工具 (wangchujiang.com)
1 | grep ^'>' 3G5U.fasta # 筛选带有 `>` 的行 |
-
PROSITE正则表达式语法: ScanProsite user manual (expasy.org)-
可通过字符串的
.replace()方法将其转换为Python的re模块的语法1
2
3
4
5
6
7
8
9
10
11
12# 此正则表达式用于寻找 钙结合的类EGF结构域签名(PROSITE ID: EGF_CA; PROSITE AC: PS01187)
pattern = '[DEQN]-x-[DEQN](2)-C-x(3,14)-C-x(3,7)\
-C-x-[DN]-x(4)-[FY]-x-C'
pattern = pattern.replace('{','[^')
pattern = pattern.replace('}',']')
pattern = pattern.replace('(','{')
pattern = pattern.replace(')','}')
pattern = pattern.replace('-','')
pattern = pattern.replace('x','.')
pattern = pattern.replace('>','$')
pattern = pattern.replace('<','^')
print pattern
-
-
在基因组序列中找到转录因子的结合位点:
转录因子结合位点列表(TFBS)作为检索表达式, 检索基因组序列即可- 转录因子: 批量预测转录因子(TF)和转录因子结合位点(TFBS) (tencent.com)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21import re
genome_seq = open('genome.txt').read()
# read transcription factor binding site patterns
sites = []
for line in open('TFBS.txt'):
fields = line.split()
tf = fields[0]
site = fields[1]
sites.append((tf, site))
# match all TF's to the genome and print matches
for tf, site in sites:
tfbs_regexp = re.compile(site)
all_matches = tfbs_regexp.findall(genome_seq)
matches = tfbs_regexp.finditer(genome_seq)
if all_matches:
print tf, ':'
for tfbs in matches:
print '\t', tfbs.group(), tfbs.start(), tfbs.end() -
从PubMed的HTML页面提取标题和摘要文本:
- 通过访问想要的网页然后使用浏览器的
开发者模式, 得到想要获取的内容的标签, 这样可以构建正则表达式来检索我们想要的内容.
- 通过访问想要的网页然后使用浏览器的
1 | import urllib2 # 网络访问库 |
- 检测科学摘要中特定的词或词组:
1 | import urllib2 |
- 氨基酸磷酸化位点:
- 简单的匹配可用正则表达式:
'R.[ST][^P]' - 参考: NetPhos 3.1–蛋白磷酸化位点预测 - 简书 (jianshu.com)
- 简单的匹配可用正则表达式:
⚓️第三部分 模块化编程
这一部分作者希望读者可以掌握编写由功能较为独立的完善的模块组成的程序
⚓️第10章 将程序划分为函数
⚓️10.2 案例: 处理三维坐标文件
-
.PDB格式解读: -
PDB-101: Learn: Guide to Understanding PDB Data: Primary Sequences and the PDB Format (rcsb.org)
- PDB文件说明 - 简书 (jianshu.com) 推荐阅读
- PDB(Protein Data Bank)数据格式详解 - Raymone1125 - 博客园 (cnblogs.com)
- 该格式文件获取来源: RCSB PDB: Homepage
(此记录为书中例子这里摘录作为一个简单的例子)
记录的第一部分此为标头, 由几个解释行组成, 包括来源生物体/实验技术(X射线/核磁共振)/交叉引用/实验细节/原始分子序列和变异残基(如果有)等. 记录的第二部分介绍了标准组的原子坐标行. 每一个原子坐标行准确地描述了一个原子. 他们以**‘ATOM’**字符串开始: 16列为’ATOM’, 711列为’原子序号’, 1316列为’原子名’, 17列’另存位置指示’, 1820列为’残基名’, 22列为’链标识符’, 2326列为’残基序号’, 27列为’残基插入码’, 3138列为x笛卡尔坐标(x轴单位为埃), 3946列为’y…‘, 4754列为’z…’, 5560列为’布局’, 6166列为’温度系数’, 7778列为’元素符号’, 7981列为’原子电荷’
(值得说明的是
原子名CA和CB指的都是碳原子,A和B用于远程标识符, 表示离氨基碳原子的远近,A: α,B: β,G: γ等,原子名称的最后一个字符可以代表分支标识符) -
struct模块: [7.3. struct — Interpret strings as packed binary data — Python 2.7.18 文档](https://docs.python.org/zh-cn/2.7/library/struct.html?highlight=struct pack#module-struct)-
struct.pack(fmt, v1, v2, …)Return a string containing the values
v1, v2, ...packed according tothe given format. The arguments must match the values required by the format exactly. -
struct.unpack(fmt, string)Unpack the string (presumably packed by
pack(fmt, ...)) according to the given format. The result is a tuple even if it contains exactly one item. The string must contain exactly the amount of data required by the format (len(string)must equalcalcsize(fmt)).
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15import struct
# pack() method; creates a string
format = '2s1s1s1s1s'
group = struct.pack(format, '10', '2', '3', '4', '5') # 结果: 102345
# unpack() method; parses the string to a tuple
format = '1s2s1s1s'
line = '12345'
col = struct.unpack(format, line) # 结果: ('1', '23', '4', '5')
# calcsize() returns the number of characters
# in a given format string
format = '30s30s20s1s'
print struct.calcsize(format) # 结果: 81 -
-
函数定义与使用: 定义函数 4. 其他流程控制工具 — Python 2.7.18 文档 详细阅读
1 | def fib(n): # write Fibonacci series up to n |
- 处理PDB数据里的坐标数据变成氨基酸序列:
1 | ''' |
-
range()迭代利器: range() 2. 内置函数 — Python 2.7.18 文档 -
可变参数与不可变参数:- 不要使用不可变参数作为函数的默认参数
-
lambda匿名函数: 4. 其他流程控制工具 — Python 2.7.18 文档
⚓️第11章 用类化繁为简
⚓️11.2 案例: 孟德尔遗传
Class(类)的构建:- 9. 类 — Python 2.7.18 文档 仔细阅读
.__repr__()类方法: 当print命令打印类时, 将按照此类方法返回的格式进行输出- 类在多人合作编程的优势
- 子类与继承
1 | class Pea: |
- 类的设计指南:
单一职责原则和不重复自己原则设计模式: Design Patterns & Refactoring (sourcemaking.com)
⚓️第12章 调试
⚓️12.2 案例: 程序无法运行时应该怎样处理
-
三种错误类型
语法错误(SyntaxError): Python规定语法之外的不合法的写法逻辑错误: 程序运行不会报错, 但是程序的运行结果不是你想要的结果运行时错误: 语法检查通过, 但是运行时出现了意想不到的情况的
-
异常处理: 8. 错误和异常 — Python 2.7.18 文档
try: ... except:语句:
1
2
3
4
5
6while True:
try:
x = int(raw_input("Please enter a number: "))
break
except ValueError:
print "Oops! That was no valid number. Try again..."
我还没有数据. 在掌握数据前建立理论是一个巨大的错误. 不知不觉地, 就开始扭曲事实以适应理论, 而不是让理论符合事实.
@ The Adventures of Sherlock Holmes, Arthur Conan Doyle
⚓️第13章 使用外部模块:R语言的Python调用接口
⚓️13.2 案例: 从文件中读取数据, 并通过Python使用R计算其平均值
R语言
⚓️第14章 构建程序流程
⚓️14.2 案例: 构建NGS流程
-
NGS技术应用:- 差异表达基因
- 非编码RNA
- 突变
-
Top Hat -
RNA-seq:
- 输出包含
小cDNA测序片段(称为读长,reads)的文件, 需回贴至参考基因组并进行拼装, 以获得全部的样本基因序列 - 某种流程实例:
- 由
Illumina(GAIIx)(或其他NGS)平台生成并存储读长至文本文件(如sample.fastq)中. TopHat可以作为RNA-seq数据的剪切接头回贴工具, 它可以把RNA-seq读长对比至哺乳动物数量级的基因组中, 参考基因组必须保存至本地以便于TopHat访问, 然后分析回贴结果, 找到外显子之间的剪切接头.TopHat读取sample.fastq得到比对读长列表, 储存在.bam文件中.Cufflinks包含有三个程序: 转录本装配(Cufflinks)/转录本比较(Cuffcompare)/检测调控和表达差异(Cuffdiff).Cufflinks进行转录本的拼接, 生成格式为transcripts.gtf的转录文件, 以便之后所用. 对于不同的样本(野生型/癌症细胞, 重复实验)还可以筛选转录组文件, 然后进行相互比较, 按照唯一的参考转录组进行拼接- 拼接好后的文件为
transcripts.gtf,可以使用Cufflink工具包中的Cuffcompare程序与其他转录组进行比较
- 由
- 输出包含
-
包装器(wrapper): 可以运行其他程序的程序 -
程序间传递数据:
raw_input()直接手动输入数据- 使用
Python变量 - 将数据存储在另一个文件里
- 使用命令行参数, 比如
sys.argv列表变量, 参见28.1. sys — 系统相关的参数和函数 — Python 2.7.18 文档
-
关闭文件的延时问题: 通过增加一个中间步骤(比如打开一个文件然后写入一些字符保存, 来确保这个中间步骤的上下两个步骤互不冲突
-
os.path操控文件目录: 10.1. os.path — 常见路径操作 — Python 2.7.18 文档 -
MSA(multiple sequence alignment, 多序列比对):Pairwise Sequence Alignment is used to identify regions of similarity that may indicate functional, structural and/or evolutionary relationships between two biological sequences (protein or nucleic acid).
By contrast, Multiple Sequence Alignment (MSA) is the alignment of three or more biological sequences of similar length. From the output of MSA applications, homology can be inferred and the evolutionary relationship between the sequences studied.
-
T-COFFEE算法: T-Coffee Home Page (tcoffee.org)T-Coffee is a multiple sequence alignment package. You can use T-Coffee to align sequences or to combine the output of your favorite alignment methods (Clustal, Mafft, Probcons, Muscle…) into one unique alignment (M-Coffee).
⚓️第15章 编写良好的程序
⚓️15.2 问题描述: 不确定性
-
《编程的准则》(Dogma of Programming):
- 著: Donald Knuth
-
软件工程周期:
- 搜集需求, 形成以用户为视角的
用户故事 - 构建简单有效的模块
- 构成整个程序, 按照新需求修改
- 搜集需求, 形成以用户为视角的
-
PEP8代码规范: -
pylint: -
代码管理工具:Git: Git手册
-
python代码自动打包工具
-
python代码生成可执行文件
py2exe: FrontPage - py2exe.orgpyinstaller: PyInstaller bundles Python applications
-
敏捷开发: 敏捷软件开发宣言 (agilemanifesto.org)
敏捷软件开发宣言
我们一直在实践中探寻更好的软件开发方法,
身体力行的同时也帮助他人。由此我们建立了如下价值观:个体和互动 高于 流程和工具
工作的软件 高于 详尽的文档
客户合作 高于 合同谈判
响应变化 高于 遵循计划也就是说,尽管右项有其价值,
我们更重视左项的价值。Kent Beck Mike Beedle Arie van Bennekum Alistair Cockburn Ward Cunningham Martin Fowler James Grenning Jim Highsmith Andrew Hunt Ron Jeffries Jon Kern Brian Marick Robert C. Martin Steve Mellor Ken Schwaber Jeff Sutherland Dave Thomas 著作权为上述作者所有,2001年
- 适于科研界的工业化实例: Helix ALM | ALM Tools | Perforce
- 对生物信息学研究的各种软件工程技术(如自动化测试/代码评审/用户故事)都有描述的工具箱: A toolbox for developing bioinformatics software | Request PDF (researchgate.net)
⚓️第四部分 数据可视化
⚓️第16章 创建科学图表
⚓️16.2 案例: 核糖体的核苷酸频率
2009年的诺贝尔化学奖授予了Venkatraman RamaKrishnan, Thomas Steitz 和 Ada Yonath, 以表彰他们对核糖体结构和功能的研究. 核糖体是已知的最大的反子机器之一, 由三种RNA组分(原核生物中是23S, 16S 和 5S rRNA) 和许多的蛋白质组成. RNA的构成包括四种基本的核糖核苷酸和一些起微调核糖体功能作用的修是核糖体
matplotlib:
1 | from pylab import figure, plot, savefig |
⚓️第17章 使用PyMOL创建分子图像
⚓️17.2 示例: 锌指
-
锌指:
锌指(英语: Zinc finger),又称锌手指, 是一种小的蛋白质结构模体, 其特征在于配合一个或多个锌离子(Zn2+)以稳定折叠. 最初用来描述非洲爪蟾卵母细胞转录因子IIIA假说结构的手指状外观
一个锌指包含由两个螺旋, 它们通过一个锌原子形成特殊的构象, 导致一个螺旋与DNA的大沟匹配
锌指名称现在已经包含了各种不同的蛋白质结构
-
PyMol: PyMOL | pymol.orgPyMOL is a user-sponsored molecular visualization system on an open-source foundation, maintained and distributed by Schrödinger.
⚓️第18章 处理图像
⚓️18.2 案例: 画一个质粒
1977年 Francisco Bolivar 和 Raymond Rodriguez 构建了最早的人工质粒. 它包含了4361个DNA碱基对, 一个复制起点, 一个氨苄青霉素抗性基因和一个四环素抗性基因. 该质粒由许多限制性位点, 从而成为构建遗传载体的基础.
-
PIL(Python Imaging Library)图形库:The Python Imaging Library (PIL) adds image processing capabilities to your Python interpreter. This library supports many file formats, and provides powerful image processing and graphics capabilities.
PIL 仅支持Python2
-
Pillow — Pillow (PIL Fork) 8.0.0 documentation
Pillow is the friendly PIL fork by Alex Clark and Contributors. PIL is the Python Imaging Library by Fredrik Lundh and Contributors.
Pillow 支持Python3
-
python-pillow/Pillow: The friendly PIL fork (Python Imaging Library) (github.com)
-
蛋白质结构域架构:
-
InterPro数据库: InterPro (ebi.ac.uk)InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites. To classify proteins in this way, InterPro uses predictive models, known as signatures, provided by several different databases (referred to as member databases) that make up the InterPro consortium. We combine protein signatures from these member databases into a single searchable resource, capitalising on their individual strengths to produce a powerful integrated database and diagnostic tool.
-
⚓️第五部分 Biopython
-
Biopython 是一个计算分子生物学模块的集合, 用它可以实现许多生物信息学项目中的基本任务
Biopython is a set of freely available tools for biological computation written in Python by an international team of developers.
-
解析生物数据格式
-
从资源库下载资源
-
运行常用的生物信息学算法
-
运行Biopython实现的算法, 进行聚类, 机器学习, 数据分析和可视化
-
各种文档索引: Documentation · Biopython
-
⚓️第19章 使用序列数据
⚓️19.2 案例: 如何将一条DNA编码序列翻译成对应的蛋白质序列并把它写入FASTA文件
Chapter 3 Sequence objects (biopython.org)
1 | from Bio.Seq import Seq |
-
IUPAC字符集:预编译好的字符集, 覆盖了所有生物序列. 包括 IUPACUnamiguousDNA(基本的ATCG字母), IUPACAmbiguousDNA(包含二义字母), ExtendedIUPACDNA(包含修饰的碱基), IUPACUnamiguousRNA, IUPACAmbiguousRNA, ExtendedIUPACRNA, IUPACProtein(IPUAC标准氨基酸)和ExtendedIUPACProtein(包括硒代半胱氨酸, X等)
-
迭代器:
一个迭代器是产生一系列的项(如SeqRecord对象)的一种数据结构. 它可以像列表一样使用在循环中, 但技术上它不是一个列表. 一个迭代器没有长度, 也不能被索引或者是切片. 用户只能向它申请下一个元素, 当你这样做时, 迭代器寻找是否存在更多的可用的记录. 这种方式下, 迭代器无需把所有的记录一致放在内存中.
-
NCBI核酸翻译编码标准: The Genetic Codes
⚓️第20章 从网络资源中检索数据
⚓️20.2 案例: 在PubMed中用关键词搜索文献, 下载并解析对应的记录
Chapter 9 Accessing NCBI’s Entrez databases (biopython.org)
⚓️第21章 使用三维结构数据
⚓️21.2 案例: 从PDB文件中提取原子名及其三维坐标
Chapter 11 Going 3D: The PDB module (biopython.org)
⚓️第六部分 编程秘笈
- 编程秘笈01: PyCogent库
PyCogent: pycogent/pycogent: PyCogent: Official repository for software and unit tests (github.com)
PyCogent includes connectors to remote databases, built-in generalized probabilistic techniques for working with biological sequences, and controllers for 3rd party applications.
项目疑似不再维护
参考文献: PyCogent: a toolkit for making sense from sequence | Genome Biology | Full Text (biomedcentral.com)
- 编程秘笈02: 反向互补和随机化序列
9.6. random — 生成伪随机数 — Python 2.7.18 文档
为什么要随机化序列:
每个序列模式可能在序列中随机出现, 在蛋白质和核酸中寻找匹配的功能模体不能保证该匹配一定有生物意义. 为了评估一个模体存在的生物学显著性, 需要检查的序列一啊不能要和随机集合进行比较. 在给定的序列集合中相对于随机序列集合有更显著表达的序列模体, 更倾向于编码一个有功能的性质(更有生物学意义). 一个好的随机序列是缺乏生物学意义的, 但是却与生物对象有相同的氨基酸/核苷酸组分, 可以通过对原序列反向,洗牌,随机化等方式创建.
1 | seq = 'ABCDEFGHIJKLMNOPQRSTUVWXYZ' |
- 编程秘笈03: 用概率创建随机序列
1 | import random |
assert 语句: assert (python.org)
assert 语句是在程序中插入调试性断言的简便方式:
1 | assert_stmt ::= "assert" expression ["," expression] |
简单形式 assert expression 等价于
1 | if __debug__: |
- 编程秘笈04: 用Biopython解析多序列联配
多序列联配(MSA):
Pfam数据库: Pfam: Home page
常用格式为.sth (Stockholm 格式)
biostar handbook(六)| 序列联配 - 简书 (jianshu.com)
例子: Pfam: Family: Globin (PF00042) (xfam.org)
通过Biopython的Bio.AlignIO模块解析多联配:
Chapter 6 Multiple Sequence Alignment objects (biopython.org)
- 编程秘笈05: 从多序列联配中计算共有序列
共有序列(Consensus Sequence): 多联配中每列中频率最高的残基(最保守的)
1 | seqs = [ |
- 编程秘笈06: 计算系统发生树的节点间的距离
Chapter 13 Phylogenetics with Bio.Phylo
Newick格式是表示系统发生树的一种统一格式. 它可以被大多数的程序读取, 包括MrBayes, GARLI, PHYLIP, TREE-PUZZLE等. Newick树包括物种和祖先节点的名字, 它们的相互关系, 以及每个节点赋予的值(通常用于父节点的距离)
- 编程秘笈07: 核苷酸序列的密码子频率
1 | # This program calculates the codon frequency in a RNA sequence |
- 编程秘笈08: 解析Vienna格式的RNA二级结构
RNA序列最重要的性质之一是其折叠和配对的能力. 与DNA不同, RNA的碱基配对不限于双螺旋, 而是能够形成复杂的结构. 预测这些RNA的碱基配对和二级结构是RNA生物信息学的常见任务.
Vienna包: ViennaRNA Web Services (univie.ac.at)
一个命令行和网络的工具集, 可以进行如从RNA序列预测碱基配对等基础任务
这些结果通常被保存为Vienna格式
1 | > two hairpin loops |
第一行包含了RNA序列名称, 第二行是序列本身
第三行的点-括号结构表达RNA二级结构
对应的括号指示一个碱基对, 而不配对的碱基对用点表示
1 | class RNAStructure: |
- 编程秘笈09: 解析BLAST的XML输出
Biopython BLAST XML解析器读取XML BLAST输出文件
7.3 Parsing BLAST output (biopython.org)
NCBIXML.parse()函数返回一个blast_out实例, 它包含一到多个记录对象(BLAST结果在一个记录里, 而如PSI-BLAST则是对每个查询有一个纪录). 每个查询都包含一到多个匹配(hit)或联配(alignment), 依次包含一到多个局部联配(HSP).
1 | blastp -query P05480.fasta -db nr.00 -out blast_output.xml |
- 编程秘笈10: 解析SBML文件
Systems Biology Markup Language (SBML), a free and open interchange format for computer models of biological processes. SBML is useful for models of metabolism, cell signaling, and more. It continues to be evolved and expanded by an international community.
SBML格式用来储存通路,反应和调控网络信息的标准格式
19.5. XML处理模块 — Python 2.7.18 文档
1 |
|
1 | from xml.dom.minidom import parse |
- 编程秘笈11: 运行BLAST
The NCBI provides a suite of command-line tools to run BLAST called BLAST+. This allows users to perform BLAST searches on their own server without size, volume and database restrictions. BLAST+ can be used with a command line so it can be integrated directly into your workflow.
Download BLAST Software and Databases Documentation (nih.gov)
7.2 Running BLAST locally (Biopython)
- 编程秘笈12: 访问、 下载和读取网页
20.5. urllib — Open arbitrary resources by URL — Python 2.7.18 文档
1 | from urllib import urlopen |
20.6. urllib2 — extensible library for opening URLs — Python 2.7.18 文档
- 编程秘笈13: 解析HTML文件
19.1. HTMLParser — Simple HTML and XHTML parser — Python 2.7.18 文档
1 | from HTMLParser import HTMLParser |
- 编程秘笈14: 将PDB文件分割成PDB链文件
1 | from struct import unpack |
也可以使用终端的>将结果导出到文件
- 编程秘笈15: 在PDB结构上找到两个最靠近的Cα原子
1 | from math import sqrt |
- 编程秘笈16: 提取两个PDB链间的界面
在一个三维结构中, 两条多肽链之间的界面被定义为满足这两个条件的所有残基对:1.两个残基分别属于不同的链;2.它们之间的距离小于某个阈值
11.2 Structure representation (biopython.org)
1 | from Bio import PDB |
- 编程秘笈17: 用Modeller建立同源模型
Modeller 是一个为建立蛋白质三维结构的同源模型设计的软件包
MODELLER is used for homology or comparative modeling of protein three-dimensional structures (1,2). The user provides an alignment of a sequence to be modeled with known related structures and MODELLER automatically calculates a model containing all non-hydrogen atoms. MODELLER implements comparative protein structure modeling by satisfaction of spatial restraints (3,4), and can perform many additional tasks, including de novo modeling of loops in protein structures, optimization of various models of protein structure with respect to a flexibly defined objective function, multiple alignment of protein sequences and/or structures, clustering, searching of sequence databases, comparison of protein structures, etc.
1 | from modeller import * |
- 编程秘笈18: 用ModeRNA分析RNA三维同源模型
ModeRNA 库: moderna · PyPI 服务维护停止
分析/操纵/建模RNA三维结构
1 | from moderna import * |
- 编程秘笈19: 从三级结构计算RNA碱基配对
RNAView: 计算一个RNA分子PDB结构中的碱基配对
NDB | Tools | Downloads (rutgers.edu)
RNA Viewer, a tool for viewing RNA 2-dimensional structure, base pairs and RNA patterns. 2D structure is fully annotated according to Leontis and Westhof (2001) RNA 7:499-512.
Yang, H., Jossinet, F., Leontis, N., Chen, L., Westbrook, J., Berman, H.M. and Westhof, E. (2003) Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res, 31, 3450-3460.
PyCogent库
1 | rnaview lehz.pdb > basepair.out |
- 编程秘笈20: 结构重叠的真实实例: 丝氨酸蛋白酶催化三分子
结构重叠: 在重叠的过程中, 两个结构之一的(靶标)被放置在一个固定的位置上,另一个(探针)被平移和旋转, 直到连个结构的均方差达到最小. 一个重叠必须要求两个集合的原子数相同才能工作, 所以在重叠两个结构之前, 需要决定每个结构上哪些原子需要重叠, 这甚至可以识整个结构的所有原子.
11.8 Accessing the Protein Data Bank (biopython.org)
11.6.6 Superimposing two structures (biopython.org)
1 | # Superimpose the catalytic triads of two different serine |
⚓️附录
⚓️附录A 命令概览
Python命令见: Python3 教程 | 菜鸟教程 (runoob.com)
⚓️附录B Python资源
Python官网: Welcome to Python.org
Python官方文档集合: 3.9.0 Documentation (python.org)
Python标准库: The Python Standard Library
Biopython教程: Biopython Tutorial and Cookbook
⚓️附录C 记录样板
1 | # C.1 A SINGLE PROTEIN SEQUENCE FILE IN FASTA FORMAT |

 是系统的
是系统的 是
是 是
是 是
是 是
是 是
是